This web page was produced as an assignment for Genetics 677, an Undergraduate course at UW-Madison
Protein-Protein Interactions
Studying the way proteins interact with other proteins is as important as studying the protein or genome itself. We study proteomics for many reasons. Proteins are the main component of physiological metabolic pathway. Giving us a better understanding of the organism, proteins differ from cell to cell and time to time while the genome does not. Furthermore, small world networks can be studied looking at primary interactions of proteins or large scale studies including secondary, tertiary or higher interactions can be organized on an Interactome map. From studying these interactions we can understand the way proteins will react to certain stresses in the environment and how these reactions will affect other proteins and relationships in cell and more importantly the human body.
STRING
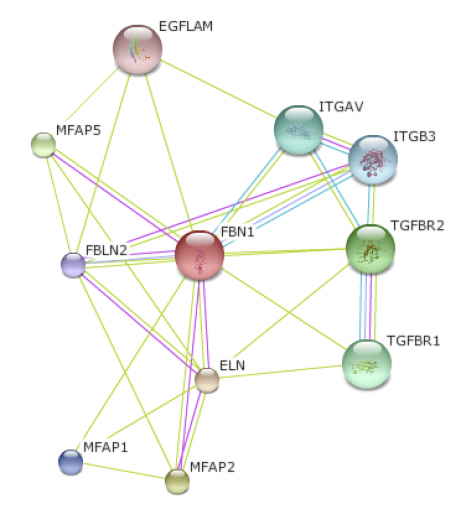
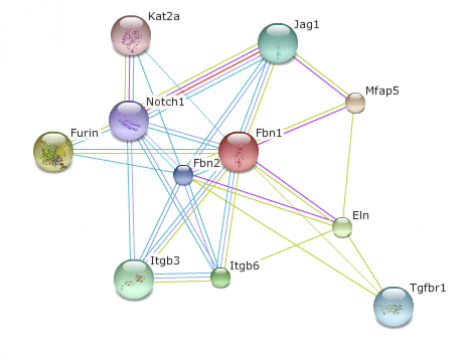
A database of both known and predicted protein interactions, STRING presents direct and indirect protein associations from various sources including high-throughput experiments to primary literature. The database provides interactive visuals of small world protein networks. By searching a protein, an interactome map will be created in various organisms. I searched for FBN1 in both humans and mice, the closest homolog on the database. By clicking on the node or edge, a description of the protein and its function will appear. Between the two interactome maps, the mouse map included proteins the human map did not. This is most likely due to the fact that more research has been done on the mouse model organism. Both maps do include the Elastin protein (ELN), which is part of the Fibrillin pathway. Elastin is also involved in aorta structure. Since problems with the aorta arise in Marfan syndrome patients, it would be beneficial to further research the interaction between Fibrillin and Elastin. (1)
Osprey and BioGRID
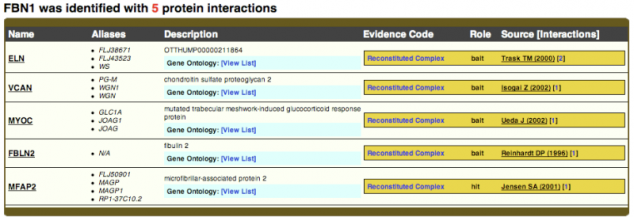
Biogrid did not have as much information as string. Only 5 proteins were found that interact with FBN1 in humans. Although FBN1 was listed to be found in other organism, the database did not have any associations listed in those organisms. Since Biogrid is the database used to create interactome maps in Osprey, the information for FBN1 is also limited in Osprey. (2) This shows that it is important to search multiple databases for information because some may have more information than others depending on the protein that is searched. BioGRID did list ELN as a protein that interacts with FBN1 as was listed in STRING. (3)
References
(1) STRING http://string.embl.de
(2) BIOGrid http://www.thebiogrid.org
(3) Osprey http://biodata.mshri.on.ca/osprey/servlet/Index
(2) BIOGrid http://www.thebiogrid.org
(3) Osprey http://biodata.mshri.on.ca/osprey/servlet/Index
Gabrielle Waclawik
[email protected]
May 13, 2010
www.gen677.weebly.com
[email protected]
May 13, 2010
www.gen677.weebly.com