This web page was produced as an assignment for Genetics 677, an Undergraduate course at UW-Madison
DNA Motifs
DNA Motifs are patterned sequences of nucleotides that can infer a biological significance such as function or process. They allow researchers to analyze sequences, identify protein families, and even develop ontologies. Motifs can be identified through a number of different algorithms. In this analysis I used MOTIF and MEME, which use the approach of deterministic optimization.
MOTIF
Using the MOTIF program, 13 motifs were identified. Rather than inputting the DNA sequence, I used the mRNA sequence because its length was not beyond the capacity of the program. I am not sure as to how this affected my search for DNA motifs. The cut-off value was set at 100. It is not known how all of these DNA motifs are involved in Fibrilllin function. I am very interested in further research of the homeo domain factor Nkx-2.5/Csx, which is a tinman homolog. Research on the tinman gene in drosophila has proven it to be essential for vertebrate heart development. Since the major cause of death in Marfan Syndrome patients has to due with the heart, I am curious if mutations in this motif can cause more severe versus mild heart problems. Other DNA motifs make no sense to me, such as the SRY motif, sed-determining region Y gene product, found only in males. Marfan Syndrome is an autosomal genetic disorder and affects women and men equally.
MEME
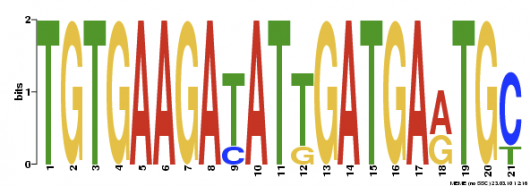
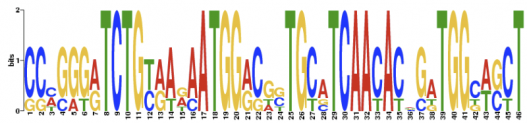
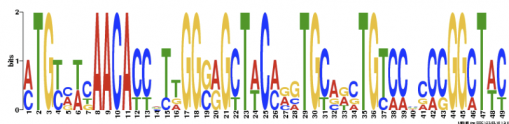
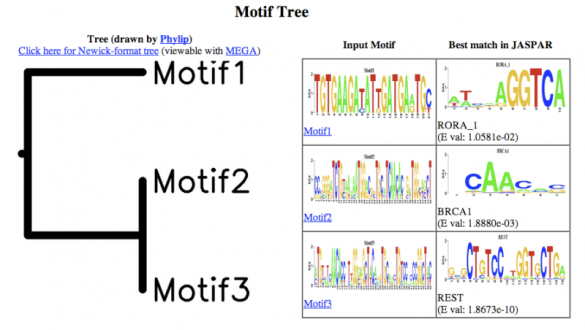
In the MEME program, three motifs were found, showing to be more stringent than MOTIF. Of the three motifs found, Motif 1 is the best conserved due to the highest P-Value calculated. You can also tell it is the best conserved just by reading the nucleotide sequence, meaning most base pair positions are only filled with a single nucleotide. No other information about the motifs was offered by MEME. I would like to further investigate this highly conserved motif by searching for similar sequences in related proteins that interact with Fibrillin-1 protein.
Motif 1:
Motif 2:
Motif 3:
Figure 2. Three DNA motifs found using MEME and their P-Value charts.
Motif Tree
References
(1) MOTIF http://motif.genome.jp/
(2) MEME http://meme.sdsc.edu/meme4/intro.html
(3) STAMP http://www.benoslab.pitt.edu/stamp/
(2) MEME http://meme.sdsc.edu/meme4/intro.html
(3) STAMP http://www.benoslab.pitt.edu/stamp/
Gabrielle Waclawik
[email protected]
May 13, 2010
www.gen677.weebly.com
[email protected]
May 13, 2010
www.gen677.weebly.com